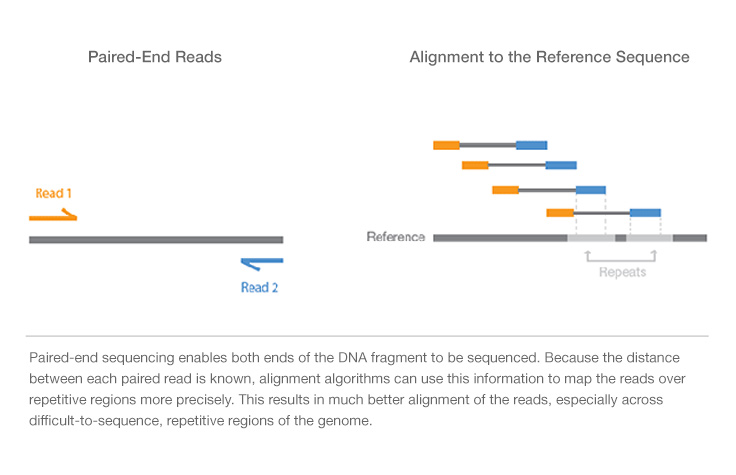
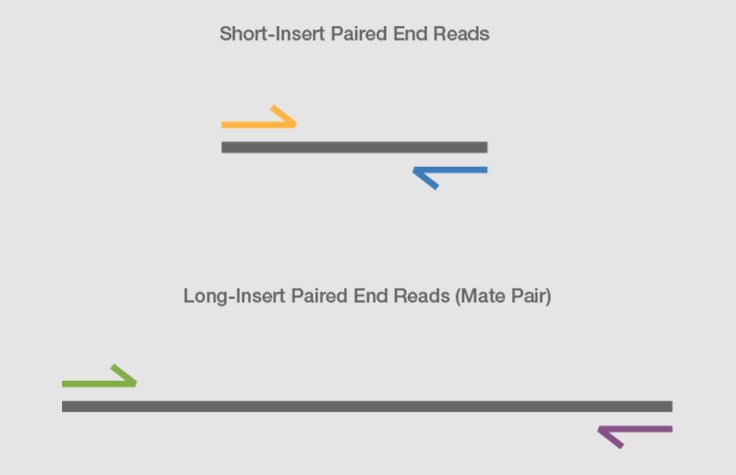
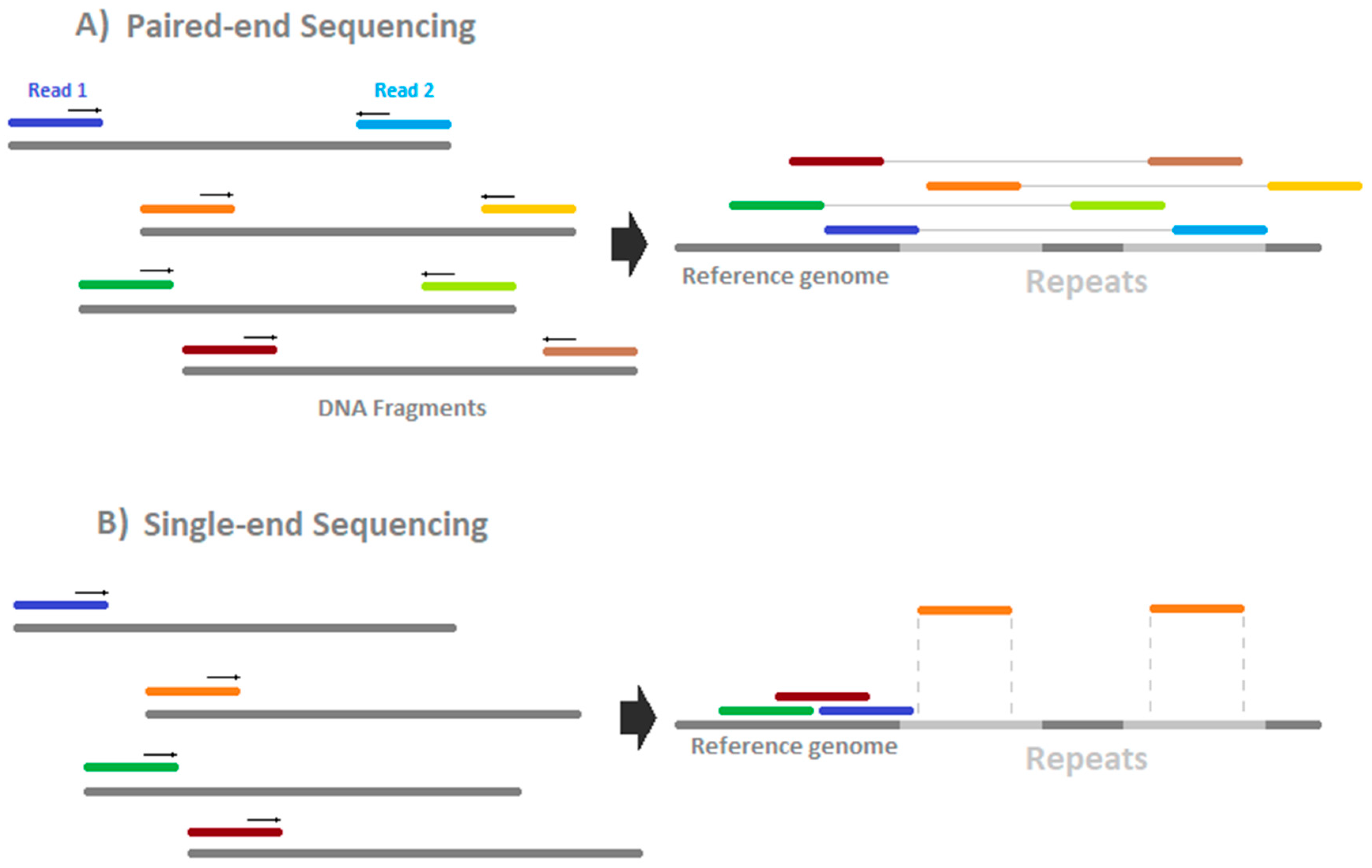
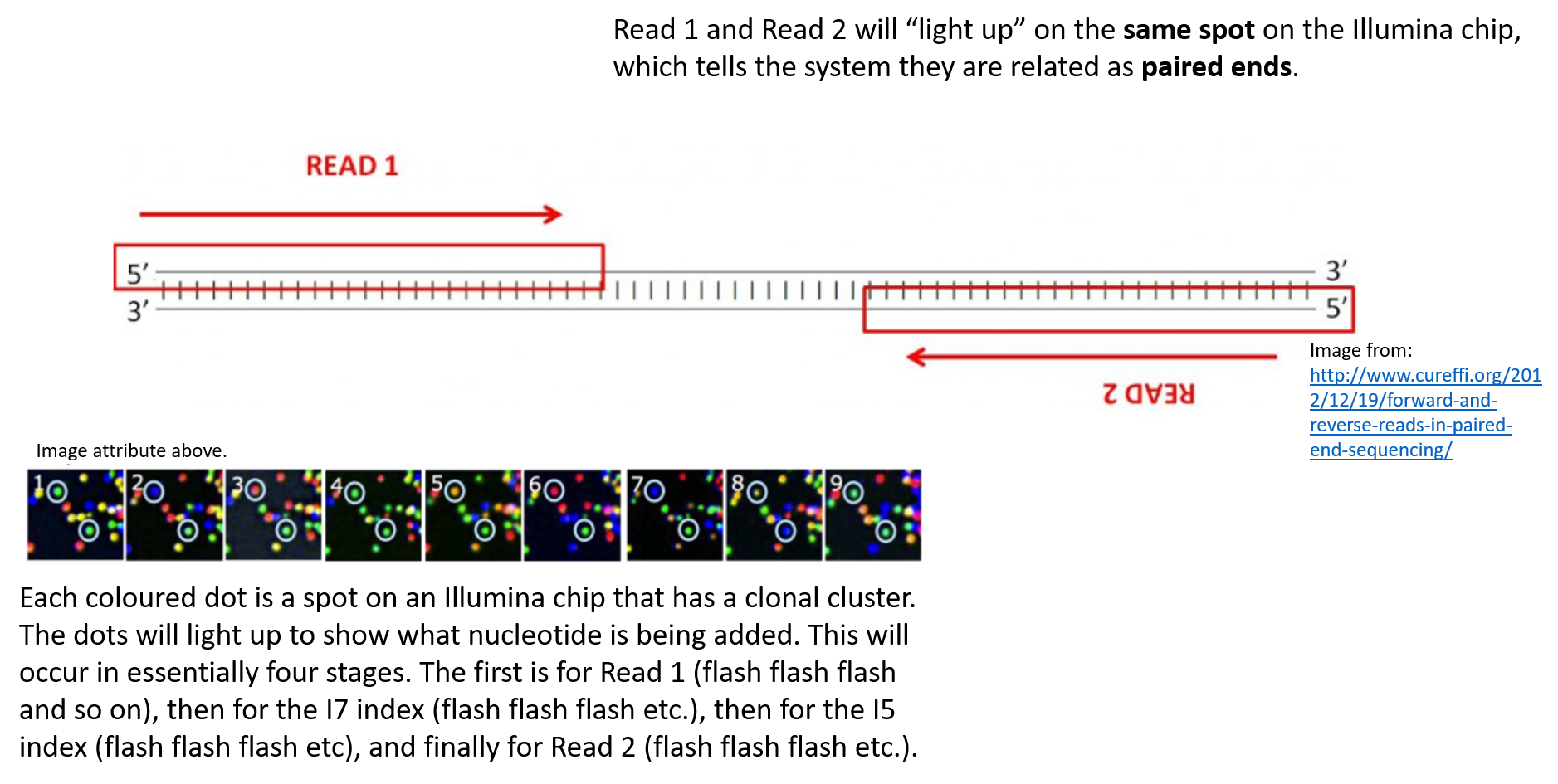
paired end sequencing advantages
Note that due to the positively skewed nature of the distribution there is a significant number of paired-end reads with a fairly long total length compared to just the individual reads themselves. FLASH to be an efficient tool in conserving reads while carrying out quality trimming in moderation.
The authors also cite the MiSeqs flexibility as an advantage a user can vary read lengths from 36 base pairs to 150 base pairs and do either single- or paired-end sequencing to enable runs to be completed in three to 27 hours.

. Mate pair sequencing is used for various applications applications including. Fast and Accurate Next-Generation Sequencing Results Enabled by Ion Torrent Technology. Next-generation Sequencing QC Standards to Enable Assay Confidence.
NGS analysis Illumina sequencing Benefits of paired end sequencing. This increase in total length is beneficial for sequence alignment algorithms de novo assembly algorithms spanning repetitive sequences and the detection. This can be very helpful e.
Swedish University of Agricultural Sciences. Anchoring one half of the pair uniquely to a single location in the genome allows mapping of the other half that is. Then the paired-end reads for species A can be used to optimise the choice of DNA sequence alignment program and parameters to align all unpaired end reads to the framework genome B thus enabling rapid and accurate construction of sequence.
As hmon stated most programs will have parameters to deal with paired-end sequencing and. Paired-end sequencing facilitates detection of genomic rearrangements and repetitive sequence elements as well as gene fusions and novel transcripts. As with everything you get what you pay for- paired end sequencing will always be the.
SNP arrays even those with millions of features provide genotypes of only a small fraction of the variants present in an individual. In addition to producing twice the number of reads for the same time and effort in library. Because PET represent connectivity between the tags the use of PET in genome re-sequencing has advantages over the use of single reads.
Which one is the best and why. Combining data from mate pair sequencing with those from short-insert paired-end reads provides increased information for maximising sequencing coverage across a genome 1. Does not require methylation of DNA or restriction digestion.
The advantage Ive seen of paired end sequencing is that in mRNA analysis when you sequence the RNA cDNA and want to map it against the reference genome you end up facing a problem which is that cDNA does not contain the introns. This application is called pairwise end sequencing known colloquially as double-barrel shotgun sequencing. Paired-end sequencing facilitates detection of genomic rearrangements and repetitive sequence elements as well as gene fusions and novel transcripts.
Therefore direct merging of the paired-end reads can prevent potential removal of informative reads that do not comply by the trimming tools strict checks. A set of paired-end reads must be obtained for species A as part of the genome sequencing project. TSS method RNA sequence data.
This allows you to get sequences of just the ends of larger pieces which means that any piece that contains an entire repetitive element may give you a pair of reads. Simple workflow allows generation of unique ranges of insert sizes. Theres also a great animation here that illustrates the concept of Illumina paired end sequencing.
Broad Range of Applications. Since paired-end reads are more likely to align to a reference the quality of the entire data set. Requires the same amount of DNA as single-read genomic DNA or cDNA sequencing.
This allows more accurate mapping particularly of repetitive regions. Paired-end sequencing allows users to sequence both ends of a fragment and generate high-quality alignable sequence data. Approximately 100-200 tumors can be sequenced at a resolution greater than 150kb when compared to sequencing an entire genome.
In paired-end reading it starts at one read finishes this direction at the specified read length and then starts another round of reading from the opposite end of the fragment. Benefits of paired end sequencing. Get 1 month free of our Silver Membership including 2 additional DNA reports.
Paired-end sequencing facilitates detection of genomic rearrangements and repetitive sequence elements as well as gene fusions and novel transcripts. Ad Access more DNA discoveries than has ever before been possible with Sequencing. One of the advantages of paired end sequencing over single end is that it doubles the amount of data.
I am partly agreed with the previous answer sure it increase the fidelity of the reads but in case of long reads the. Next generation sequencing-based approaches for transcription start site mapping. Ad Enable a Range of Targeted Next-Generation Sequencing Applications wSpeed and Scalability.
In addition paired-end sequencing partly alleviates the difficulty of aligning single short reads to repeat regions and thus allows a subset of repeat elements to be at least partially characterized by RNA-seq. In addition to producing twice the number of reads for the same time and effort in library. What do the numbers in BLOSSUM PAM represent.
Another supposed advantage is that it leads to more accurate reads because if say Read 1 see picture below maps to two different regions of the genome Read 2 can be used to help determine which one of the two regions makes more sense. Whats are the advantages and disadvantages of of both the process. Paired-end sequencing allows users to sequence both ends of a fragment and generate high-quality alignable sequence data.
By clicking Accept All you consent to the use of ALL the cookies. For your De novo genome assembly Fig. We use cookies on our website to give you the most relevant experience by remembering your preferences and repeat visits.
Can be used for. This means your two reads are the reverse complement of the 100 3-most bases of the Watson strand and the Crick strand. A sequencing run on the PGM meantime lasts around two hours for a 200 base run.
Paired-end sequencing reads from both ends of a DNA fragment and is capable of pairing ends together -- so you know whats on the ends of your fragments even if each individual read doesnt overlap with its mate. Because ESP only looks at short paired-end sequences it has the advantage of providing useful information genome-wide without the need for large-scale sequencing. Overall our results show that merging paired-end reads of eDNA data before trimming.
Ad Next-generation Sequencing Service Computational Analyse for PreventionEarly Detection. Paired-end sequencing allows users to sequence both ends of a fragment and generate high-quality alignable sequence data. Benefits of paired end sequencing.
Paired-end reading improves the ability to identify the relative positions of various reads in the genome making it much more effective than single-end reading in resolving structural rearrangements such as. The grey represents the fragment and each end of the fragment is sequenced. Sequencing and genome assembly o Advantages of paired end sequencing o Graph representations for genome assembly Know how to build a deBruijn from reads Alignment o Dynamical programing for global and local alignments o Scoring matricies Log-odds scoring.

End Sequence Profiling Wikiwand

Understanding The Characteristics Of Sequence Based Single Source Dna Profiles Forensic Science International Genetics

Human Genome Project Sequencing The Human Genome Learn Science At Scitable
Epigenomes Free Full Text How To Design A Whole Genome Bisulfite Sequencing Experiment Html
Illumina Sequencing For Dummies An Overview On How Our Samples Are Sequenced Kscbioinformatics

The Variables For Ngs Experiments Coverage Read Length Multiplexing

Study Of Acute Liver Failure In Children Using Next Generation Sequencing Technology The Journal Of Pediatrics

Results From Illumina Miseq Paired End Sequencing Run 2 150 Bp Download Table

Gbs Adapters Pcr And Sequencing Primers A Sequences Of Download Scientific Diagram

What Are Paired End Reads The Sequencing Center
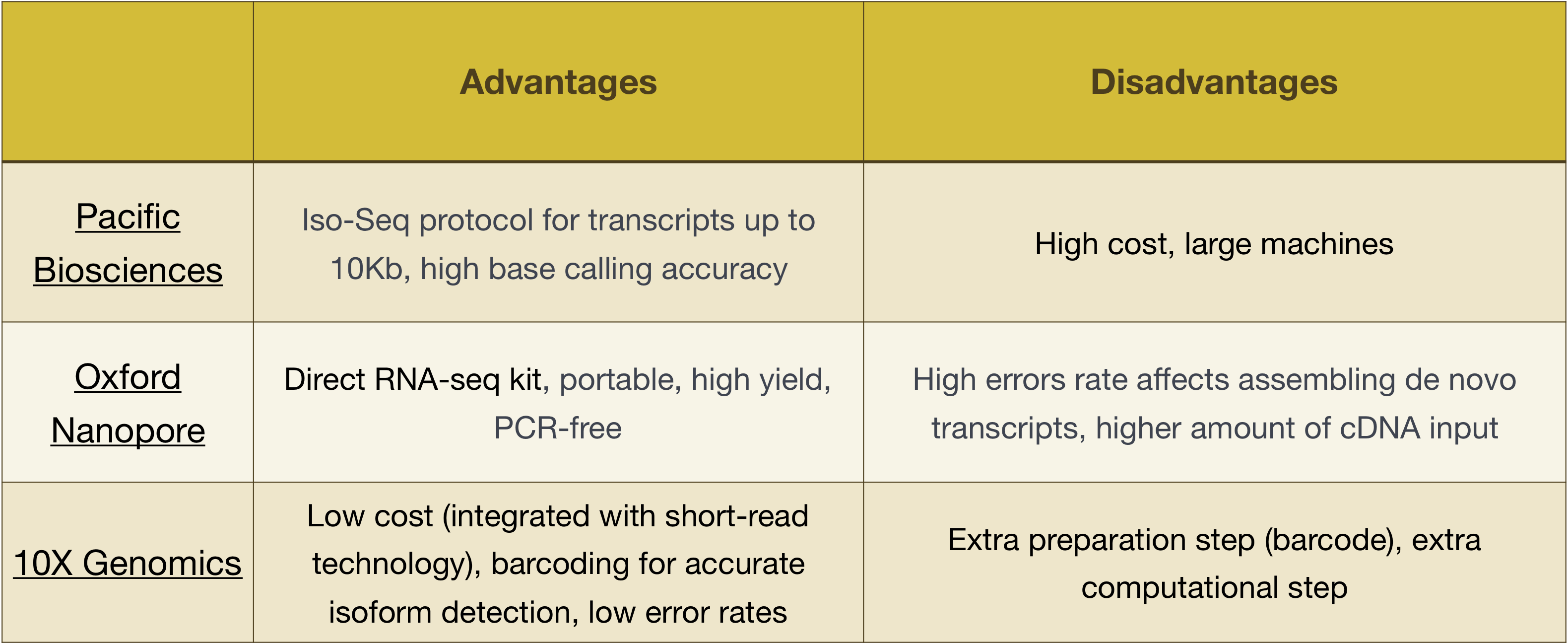
Intro To Rna Seq Introduction To Rna Seq Using High Performance Computing

Detct A Purely Quantitative Digital Gene Expression Sample Processing And Analysis Package Researchers At The We Gene Expression Analysis Sequence Analysis
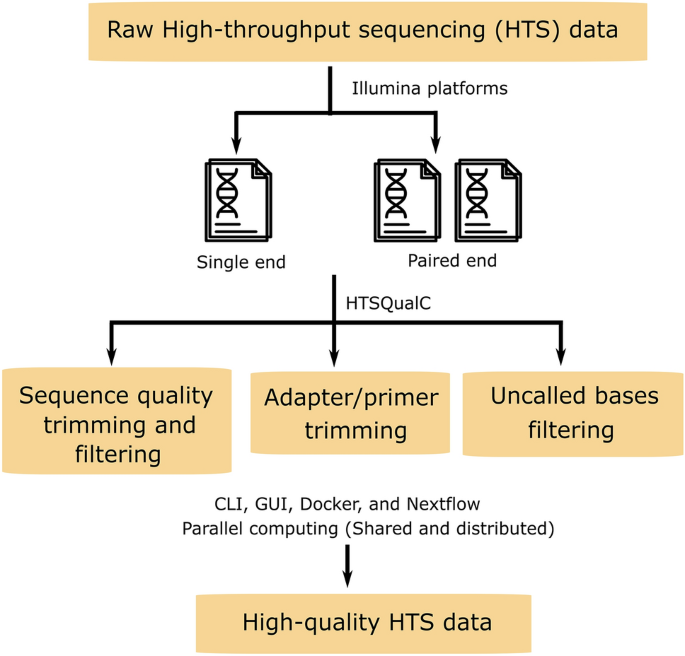
Htsqualc Is A Flexible And One Step Quality Control Software For High Throughput Sequencing Data Analysis Scientific Reports

What Is Mate Pair Sequencing For

Beyond The Linear Genome Paired End Sequencing As A Biophysical Tool Trends In Cell Biology

Detct A Purely Quantitative Digital Gene Expression Sample Processing And Analysis Package Researchers At The We Gene Expression Analysis Sequence Analysis